
[ad_1]
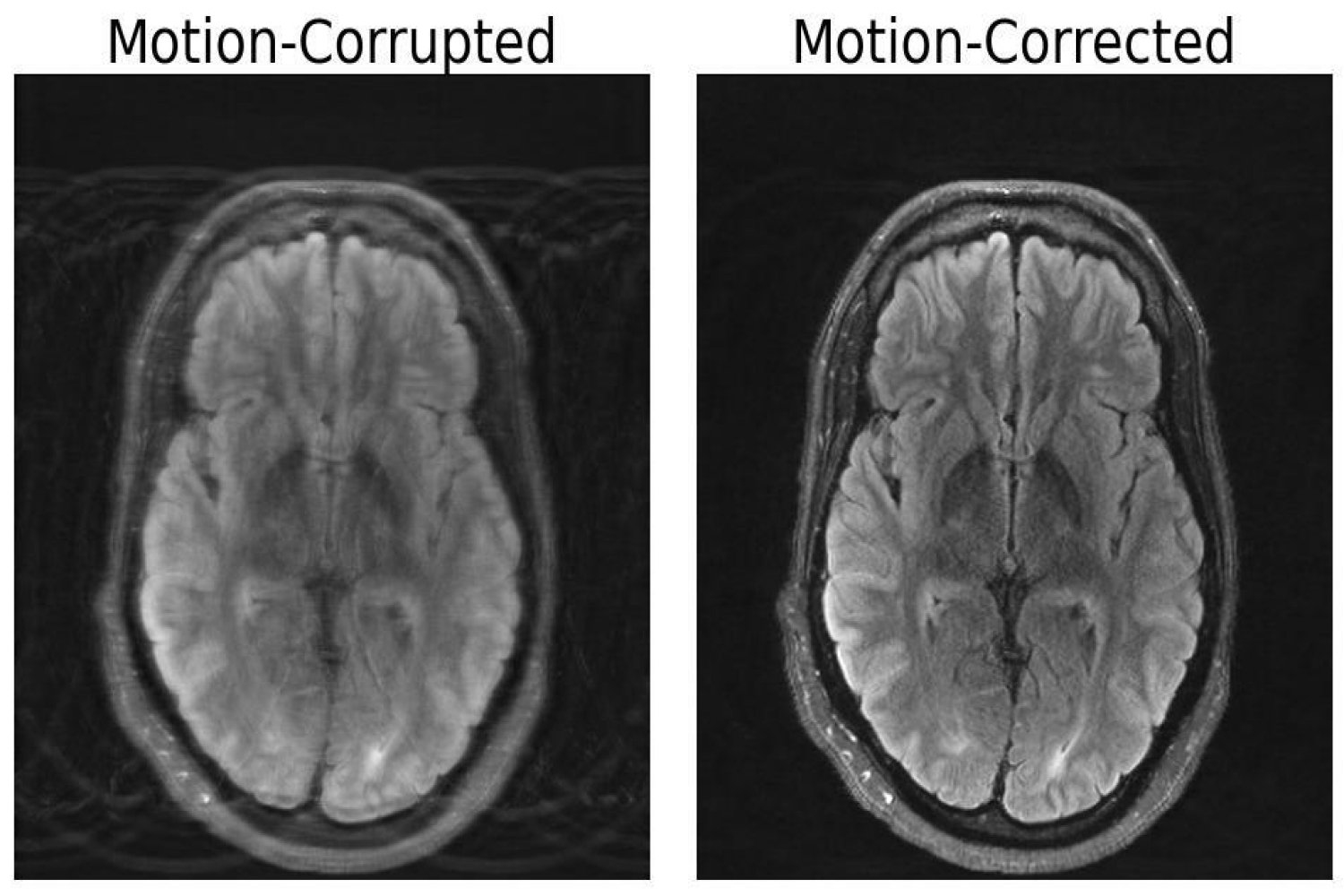
In comparison with different imaging modalities like X-rays or CT scans, MRI scans present high-quality gentle tissue distinction. Sadly, MRI is extremely delicate to movement, with even the smallest of actions leading to picture artifacts. These artifacts put sufferers prone to misdiagnoses or inappropriate therapy when essential particulars are obscured from the doctor. However researchers at MIT might have developed a deep studying mannequin able to movement correction in mind MRI.
“Movement is a standard drawback in MRI,” explains Nalini Singh, an Abdul Latif Jameel Clinic for Machine Studying in Well being (Jameel Clinic)-affiliated PhD pupil within the Harvard-MIT Program in Well being Sciences and Know-how (HST) and lead writer of the paper. “It’s a fairly gradual imaging modality.”
MRI periods can take wherever from a couple of minutes to an hour, relying on the kind of photos required. Even throughout the shortest scans, small actions can have dramatic results on the ensuing picture. Not like digital camera imaging, the place movement sometimes manifests as a localized blur, movement in MRI usually leads to artifacts that may corrupt the entire picture. Sufferers could also be anesthetized or requested to restrict deep respiration to be able to decrease movement. Nevertheless, these measures usually can’t be taken in populations notably vulnerable to movement, together with kids and sufferers with psychiatric issues.
The paper, titled “Information Constant Deep Inflexible MRI Movement Correction,” was not too long ago awarded finest oral presentation on the Medical Imaging with Deep Studying convention (MIDL) in Nashville, Tennessee. The strategy computationally constructs a motion-free picture from motion-corrupted information with out altering something concerning the scanning process. “Our purpose was to mix physics-based modeling and deep studying to get the very best of each worlds,” Singh says.
The significance of this mixed method lies inside making certain consistency between the picture output and the precise measurements of what’s being depicted, in any other case the mannequin creates “hallucinations” — photos that seem lifelike, however are bodily and spatially inaccurate, probably worsening outcomes with regards to diagnoses.
Procuring an MRI freed from movement artifacts, notably from sufferers with neurological issues that trigger involuntary motion, comparable to Alzheimer’s or Parkinson’s illness, would profit extra than simply affected person outcomes. A examine from the College of Washington Division of Radiology estimated that movement impacts 15 p.c of mind MRIs. Movement in all sorts of MRI that results in repeated scans or imaging periods to acquire photos with ample high quality for analysis leads to roughly $115,000 in hospital expenditures per scanner on an annual foundation.
In keeping with Singh, future work may discover extra subtle sorts of head movement in addition to movement in different physique elements. As an illustration, fetal MRI suffers from speedy, unpredictable movement that can not be modeled solely by easy translations and rotations.
“This line of labor from Singh and firm is the subsequent step in MRI movement correction. Not solely is it glorious analysis work, however I imagine these strategies shall be utilized in all types of medical circumstances: kids and older of us who cannot sit nonetheless within the scanner, pathologies which induce movement, research of shifting tissue, even wholesome sufferers will transfer within the magnet,” says Daniel Moyer, an assistant professor at Vanderbilt College. “Sooner or later, I feel that it probably shall be normal observe to course of photos with one thing instantly descended from this analysis.”
Co-authors of this paper embrace Nalini Singh, Neel Dey, Malte Hoffmann, Bruce Fischl, Elfar Adalsteinsson, Robert Frost, Adrian Dalca and Polina Golland. This analysis was supported partly by GE Healthcare and by computational {hardware} offered by the Massachusetts Life Sciences Middle. The analysis workforce thanks Steve Cauley for useful discussions. Further assist was offered by NIH NIBIB, NIA, NIMH, NINDS, the Blueprint for Neuroscience Analysis, a part of the multi-institutional Human Connectome Venture, the BRAIN Initiative Cell Census Community, and a Google PhD Fellowship.
[ad_2]